Inflammatory Bowel Disease (IBD), which includes Crohn’s disease and Ulcerative Colitis, affects millions of people worldwide. Although these conditions differ in the part of the digestive tract they affect, they share common features—chronic inflammation, immune dysregulation, and disruption of the gut barrier. Understanding the proteins driving these processes is essential for developing more specific and predictive biomarkers and identifying therapeutic targets.
The Challenge: Finding Disease-Specific Biomarkers
Advancing IBD biomarkers and therapeutic targets remains challenging despite the wealth of public and proprietary data. Three core challenges are particularly relevant:
Limited specificity of existing markers
Current clinical markers, such as CRP or fecal calprotectin, are sensitive but rise in many inflammatory conditions, making it difficult to identify IBD-specific signals.
Fragmented evidence
Proteins and genetic associations are reported across diverse studies, cohorts, and omics layers. Aggregating these signals into a coherent, actionable picture is time-consuming and prone to gaps. Making it hard to identify the most promising targets efficiently.
High bar for translational relevance
Biomarkers and therapeutic targets must combine sensitivity, specificity, predictive value, and clinical actionability. As a result, selecting and prioritizing them requires careful evaluation to guide experimental validation or drug development efforts.
22 Proteins Linked to IBD: Insights from a Large-Scale Study
A recent study of 48,800 participants in the UK Biobank applied multiple approaches. These included prospective cohort analysis, Mendelian Randomization, and Bayesian colocalization to identify proteins with potential causal roles in IBD. Identifying which proteins drive disease can help biotech and pharma teams prioritize the most promising biomarkers and therapeutic targets.
The researchers of the study organized the 22 proteins into two evidence tiers, based on the strength of causal links to IBD.
Tier 1: High-Confidence IBD Proteins (8 proteins)
IL12B, CD6, MXRA8, CXCL9, IFNG, CCN3, RSPO3, IL18 – proteins are supported by Mendelian randomization and colocalization analysis, meaning they share genetic variants that influence both protein levels and IBD risk.
Tier 2: Emerging or Context-Specific Candidates (14 proteins)
CCL20, TNFRSF9, CCL7, IL1RL1, IL19, CCL13, CD72, EPHB4, SEPTIN8, LY9, IL10RA, IGLC2, ITGAV, and NOS3 -proteins that are supported by Mendelian Randomization only, meaning that they are promising candidates, but further analyses are needed to confirm their direct mechanistic role in IBD.
Overall, these 22 proteins span key processes in IBD, including immune activation, chemokine signaling, epithelial repair, and vascular responses, highlighting the diverse biological mechanisms contributing to disease.
Explore these proteins in DISGENET
Using DISGENET to Contextualize IBD Proteins
With 22 candidate proteins for IBD, teams need to determine which are well-supported by evidence and ready for translational exploration, and which represent novel opportunities for early-stage discovery.
To give us a more complete picture, we mapped these proteins in DISGENET, a comprehensive genotype-phenotype platform, integrating evidence from specialized and the latest scientific literature.
Key findings from DISGENET:
- 17 of 22 proteins were already known to be associated with IBD, Ulcerative Colitis, or Crohn’s Disease before this study
- The remaining proteins link to broader immune or inflammatory conditions, highlighting potential novel biology.
Interestingly, one Tier 2 protein, TNFRSF9, reported in the study as lacking direct evidence for IBD, is in fact associated with colitis in DISGENET, with multiple supporting studies. This highlights the value of integrated resources that consolidate molecular evidence across datasets.
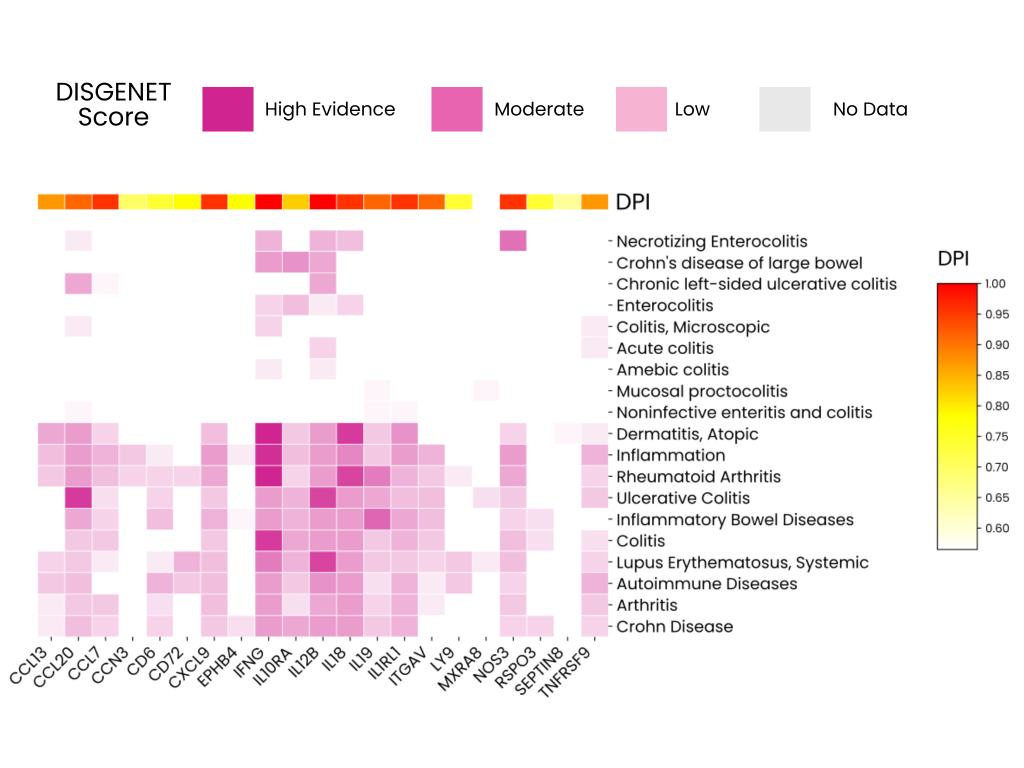
This heatmap provides an overview of how strongly each protein is linked to IBD. It also shows which proteins are more broadly inflammatory and which may represent emerging opportunities. These patterns are summarized using two key metrics:
- DISGENET Score: Measures the strength of associations based on quality and the quantity of evidence available.
- Disease Pleiotropy Index (DPI): Indicates if a protein’s effects stay within a single disease category or spread across completely different disease classes.
Biotech and Pharma Applications
For R&D teams, the value of these findings lies in identifying proteins that can move efficiently from discovery to validation and therapeutic exploration. The combination of genetic evidence, tiered protein classification, and DISGENET context provides a framework for decision-making, so biopharma teams can:
Focus on High-Confidence Therapeutic Targets
The tier 1 proteins from the study show strong causal links to IBD. These proteins are prime candidates for target validation or pathway-focused studies. By leveraging DISGENET metrics, teams can see which proteins are specific to IBD versus broadly inflammatory, helping focus resources on pathways most likely to yield disease-specific effects.
Identify Biomarkers with Maximum Potential
Evidence tiers combined with the DISGENET Score and Disease Pleiotropy Index (DPI) help identify proteins that are both biologically meaningful and IBD-specific. These insights guide early translational studies, such as patient stratification, early detection models, or monitoring treatment response. This allows teams to focus on proteins with the highest potential impact.
Spot Emerging Targets Before Competitors
Tier 2 proteins and those with broader pleiotropy may reveal emerging biology worth exploring. Using DISGENET to contextualize these candidates against existing disease associations helps de-risk early-stage studies and focus on proteins with translational potential.
Streamline Target Prioritization for Efficient Validation
Integrating genetic support, evidence tiers, and disease associations provides a systematic approach to prioritization. Proteins can be ranked by causal evidence, disease specificity, and novelty, helping teams allocate experimental resources efficiently while keeping exploratory targets in view for follow-up studies.
Curious how your team can prioritize IBD proteins more efficiently? Book a demo and explore them in DISGENET
Advancing IBD Research
Contextualizing IBD-associated proteins through integrated data layers allows biotech and pharma teams to distinguish well-established targets from emerging opportunities. Leveraging this framework helps focus R&D efforts on proteins most likely to yield high-value insights, streamline experimental validation, and maintain a competitive edge in biomarker and therapeutic discovery.
Key Takeaways
- 17 of 22 proteins are already linked to IBD, UC, or Crohn’s Disease, confirming strong biological relevance.
- Tier 1 proteins (e.g., IL12B, IFNG, CXCL9) represent high-confidence targets for biomarker validation or therapeutic exploration.
- Tier 2 proteins may reveal emerging biology, offering opportunities for early-stage discovery and novel mechanistic insights.
- DISGENET metrics (DISGENET Score and Disease Pleiotropy Index) allow teams to assess disease specificity and potential impact, streamlining prioritization decisions.
